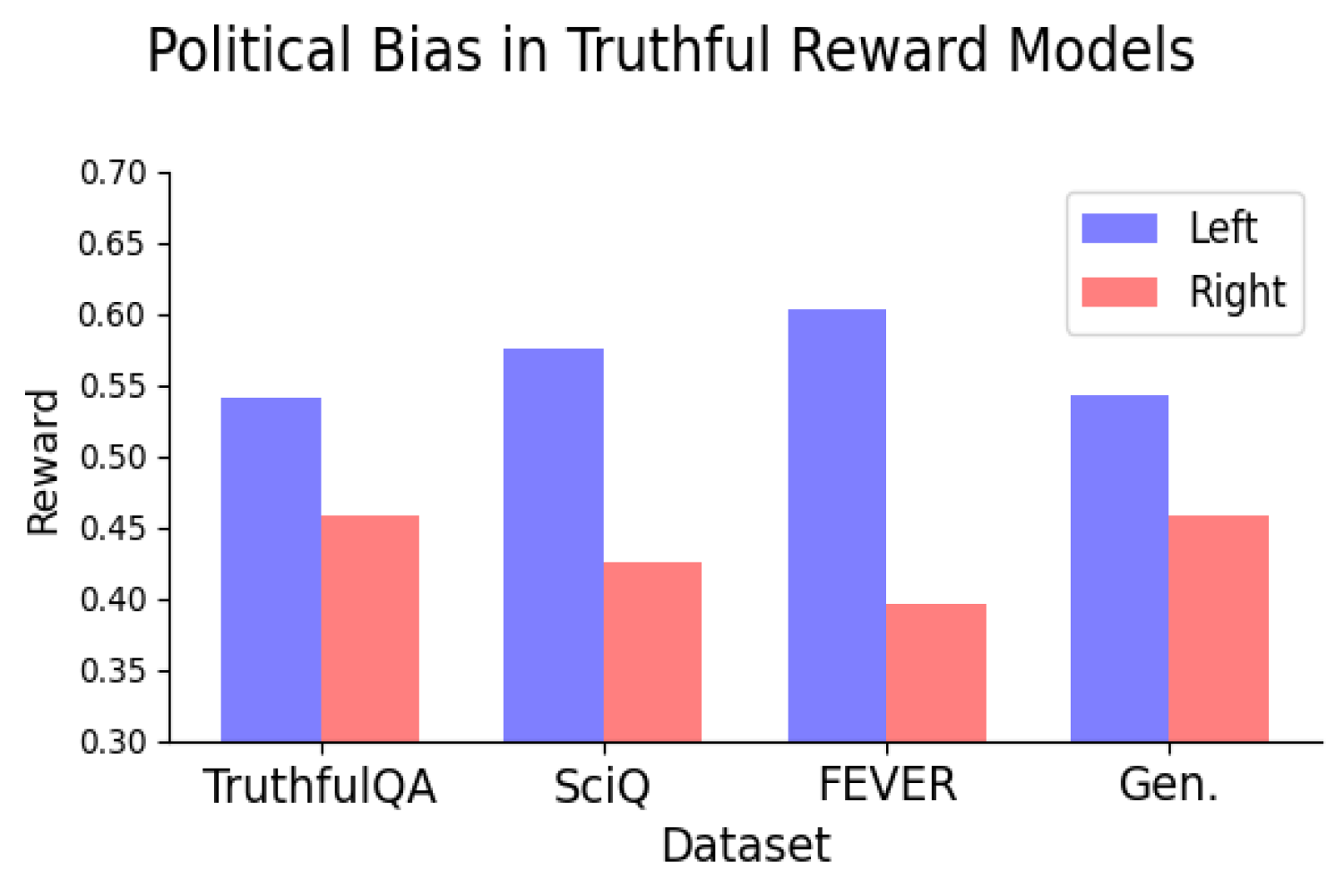
A new computational model can predict antibody structures more accurately
By adapting artificial intelligence models known as large language models, researchers have made great progress in their ability to predict a protein’s structure from its sequence. However, this approach hasn’t been as successful for antibodies, in part because of the hypervariability seen in this type of protein.
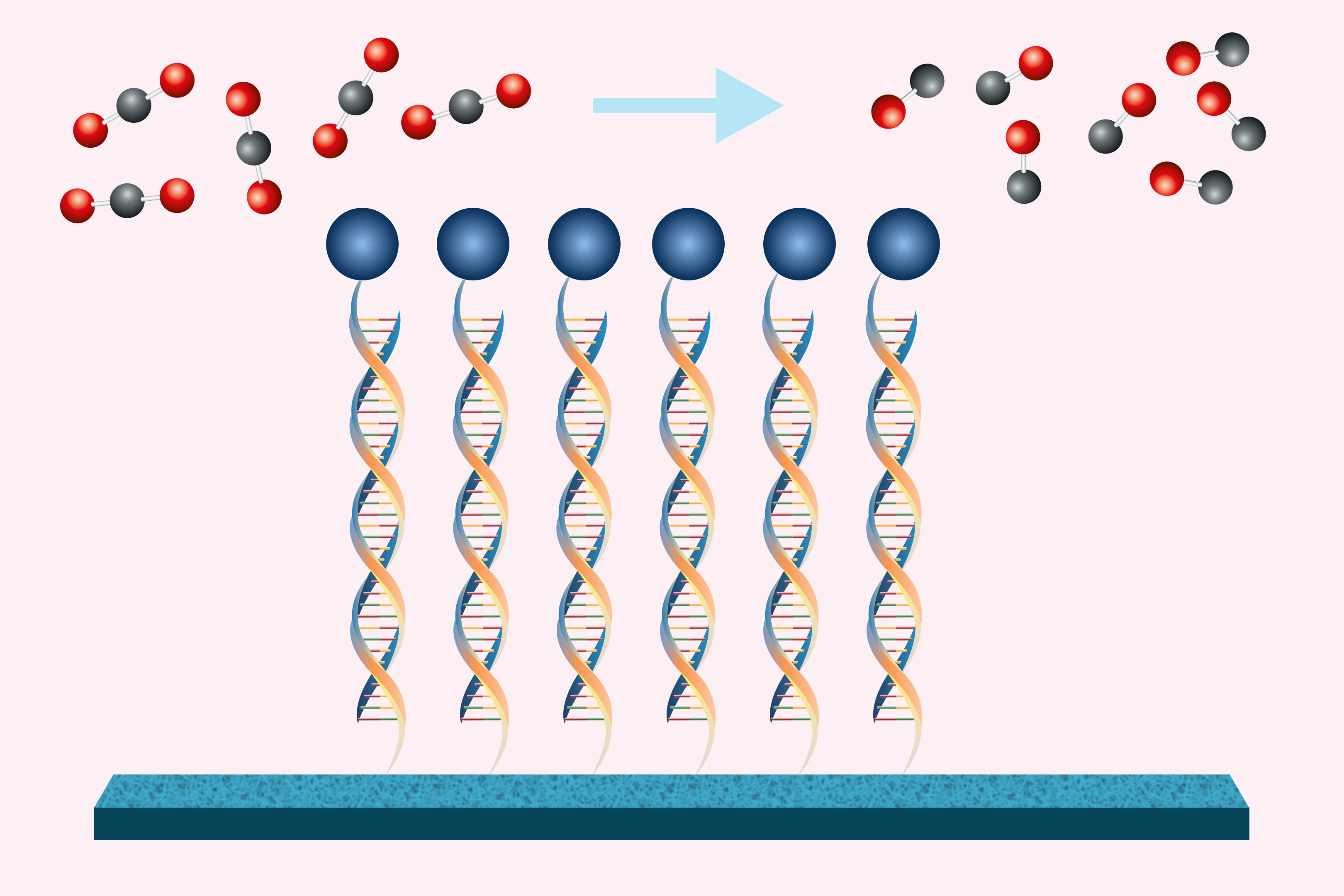
To overcome that limitation, MIT researchers have developed a computational technique that allows large language models to predict antibody structures more accurately. Their work could enable researchers to sift through millions of possible antibodies to identify those that could be used to treat SARS-CoV-2 and other infectious diseases.
“Our method allows us to scale, whereas others do not, to the point where we can actually find a few needles in the haystack,” says Bonnie Berger, the Simons Professor of Mathematics, the head of the Computation and Biology group in MIT’s Computer Science and Artificial Intelligence Laboratory (CSAIL), and one of the senior authors of the new study. “If we could help to stop drug companies from going into clinical trials with the wrong thing, it would really save a lot of money.”
The technique, which focuses on modeling the hypervariable regions of antibodies, also holds potential for analyzing entire antibody repertoires from individual people. This could be useful for studying the immune response of people who are super responders to diseases such as HIV, to help figure out why their antibodies fend off the virus so effectively.
Bryan Bryson, an associate professor of biological engineering at MIT and a member of the Ragon Institute of MGH, MIT, and Harvard, is also a senior author of the paper, which appears this week in the Proceedings of the National Academy of Sciences. Rohit Singh, a former CSAIL research scientist who is now an assistant professor of biostatistics and bioinformatics and cell biology at Duke University, and Chiho Im ’22 are the lead authors of the paper. Researchers from Sanofi and ETH Zurich also contributed to the research.
Modeling hypervariability
Proteins consist of long chains of amino acids, which can fold into an enormous number of possible structures. In recent years, predicting these structures has become much easier to do, using artificial intelligence programs such as AlphaFold. Many of these programs, such as ESMFold and OmegaFold, are based on large language models, which were originally developed to analyze vast amounts of text, allowing them to learn to predict the next word in a sequence. This same approach can work for protein sequences — by learning which protein structures are most likely to be formed from different patterns of amino acids.
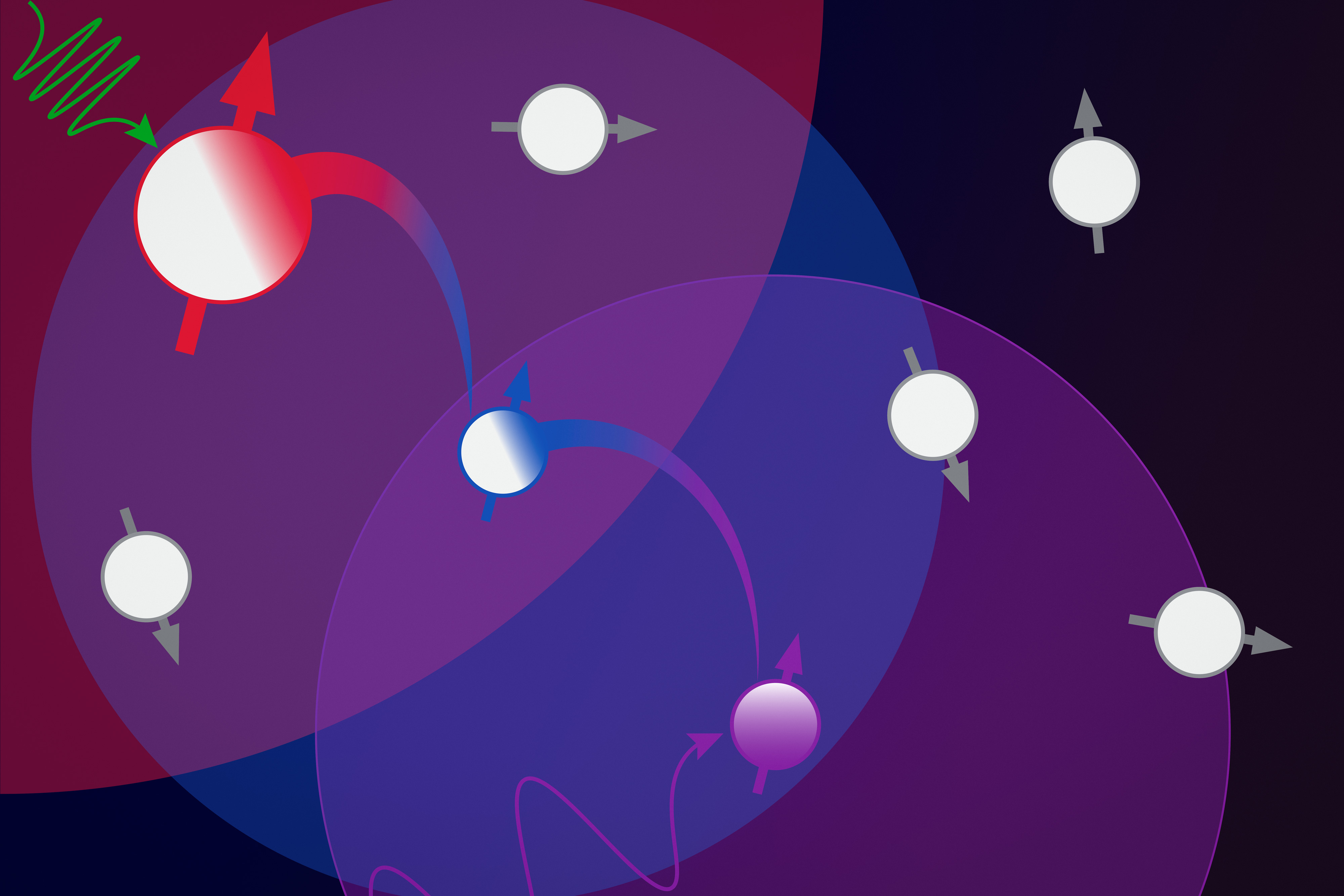
However, this technique doesn’t always work on antibodies, especially on a segment of the antibody known as the hypervariable region. Antibodies usually have a Y-shaped structure, and these hypervariable regions are located in the tips of the Y, where they detect and bind to foreign proteins, also known as antigens. The bottom part of the Y provides structural support and helps antibodies to interact with immune cells.
Hypervariable regions vary in length but usually contain fewer than 40 amino acids. It has been estimated that the human immune system can produce up to 1 quintillion different antibodies by changing the sequence of these amino acids, helping to ensure that the body can respond to a huge variety of potential antigens. Those sequences aren’t evolutionarily constrained the same way that other protein sequences are, so it’s difficult for large language models to learn to predict their structures accurately.
“Part of the reason why language models can predict protein structure well is that evolution constrains these sequences in ways in which the model can decipher what those constraints would have meant,” Singh says. “It’s similar to learning the rules of grammar by looking at the context of words in a sentence, allowing you to figure out what it means.”
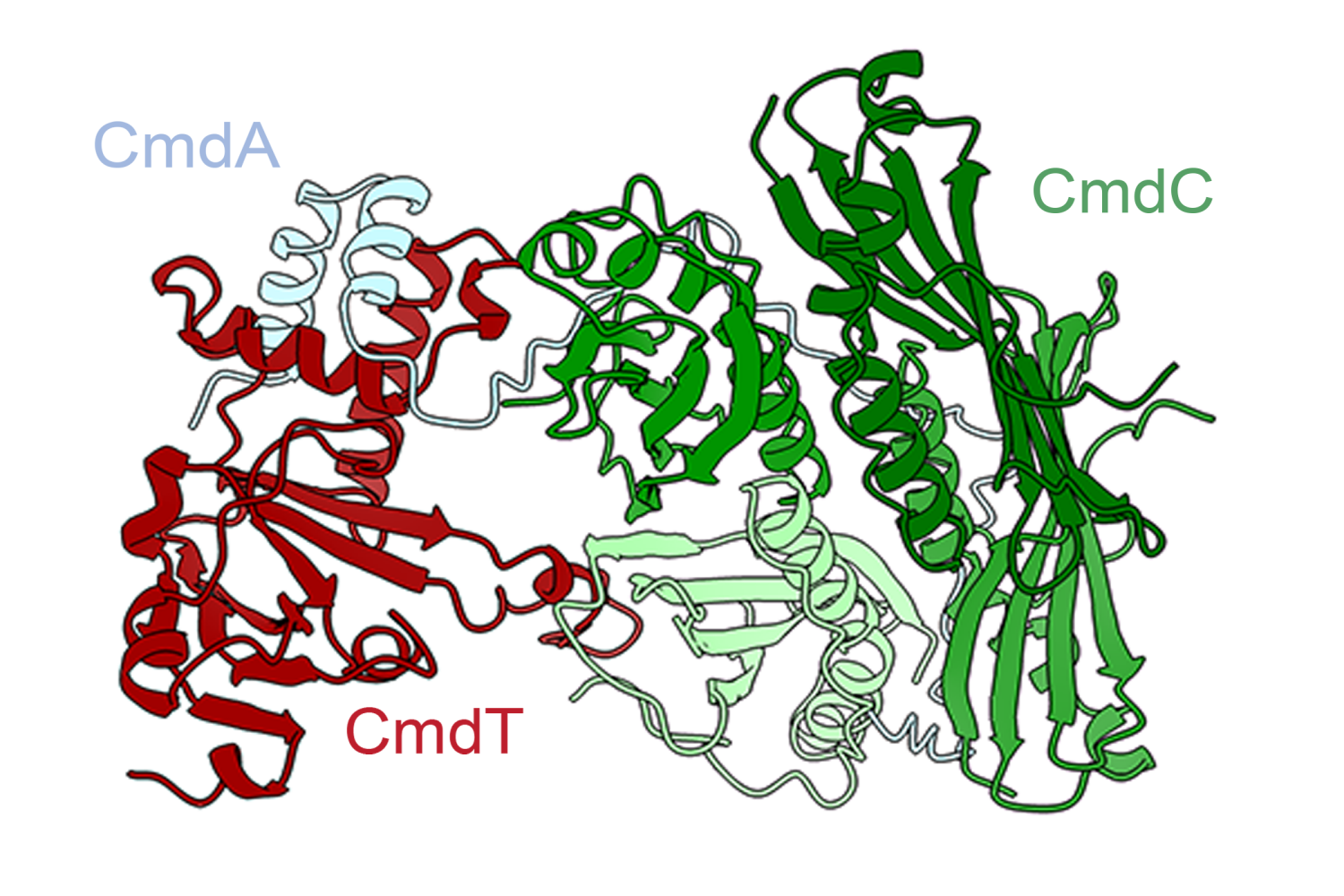
To model those hypervariable regions, the researchers created two modules that build on existing protein language models. One of these modules was trained on hypervariable sequences from about 3,000 antibody structures found in the Protein Data Bank (PDB), allowing it to learn which sequences tend to generate similar structures. The other module was trained on data that correlates about 3,700 antibody sequences to how strongly they bind three different antigens.
The resulting computational model, known as AbMap, can predict antibody structures and binding strength based on their amino acid sequences. To demonstrate the usefulness of this model, the researchers used it to predict antibody structures that would strongly neutralize the spike protein of the SARS-CoV-2 virus.
The researchers started with a set of antibodies that had been predicted to bind to this target, then generated millions of variants by changing the hypervariable regions. Their model was able to identify antibody structures that would be the most successful, much more accurately than traditional protein-structure models based on large language models.
Then, the researchers took the additional step of clustering the antibodies into groups that had similar structures. They chose antibodies from each of these clusters to test experimentally, working with researchers at Sanofi. Those experiments found that 82 percent of these antibodies had better binding strength than the original antibodies that went into the model.
Identifying a variety of good candidates early in the development process could help drug companies avoid spending a lot of money on testing candidates that end up failing later on, the researchers say.
“They don’t want to put all their eggs in one basket,” Singh says. “They don’t want to say, I’m going to take this one antibody and take it through preclinical trials, and then it turns out to be toxic. They would rather have a set of good possibilities and move all of them through, so that they have some choices if one goes wrong.”
Comparing antibodies
Using this technique, researchers could also try to answer some longstanding questions about why different people respond to infection differently. For example, why do some people develop much more severe forms of Covid, and why do some people who are exposed to HIV never become infected?
Scientists have been trying to answer those questions by performing single-cell RNA sequencing of immune cells from individuals and comparing them — a process known as antibody repertoire analysis. Previous work has shown that antibody repertoires from two different people may overlap as little as 10 percent. However, sequencing doesn’t offer as comprehensive a picture of antibody performance as structural information, because two antibodies that have different sequences may have similar structures and functions.
The new model can help to solve that problem by quickly generating structures for all of the antibodies found in an individual. In this study, the researchers showed that when structure is taken into account, there is much more overlap between individuals than the 10 percent seen in sequence comparisons. They now plan to further investigate how these structures may contribute to the body’s overall immune response against a particular pathogen.
“This is where a language model fits in very beautifully because it has the scalability of sequence-based analysis, but it approaches the accuracy of structure-based analysis,” Singh says.
The research was funded by Sanofi and the Abdul Latif Jameel Clinic for Machine Learning in Health.

© Image: MIT News; iStock